Basic Enzymology, Allostery & Drug Design
If you are interested in joining projects in any of these areas please contact Ellis Bell (jbell@sandiego.edu)
Current CUREs using Watermelon Glyoxysomal Malate Dehydrogenase are focused around six project areas:
Active Site (Catalysis)
involves residues directly or indirectly responsibe for the hydride transfer and proton abstraction steps in the overall catalysis. While clear roles for several “first sphere” catalytic residues are well established, how “second sphere” residues contribute to either catalysis of allosteric interactions are unknown.
2. Mobile Loop (Substrate Specificity etc) involves the mobile loop that closes over the active site. This loop not only controls substrate specificity (choice of 3, 4 or 5 carbon substrates) but is also thought to contribute to both catalysis and cofactor binding. While some of the factors controlling 3 carbon versus 4 carbon substrates have been explored, little is know about how the active sites accommodates the 5 carbon oncometabolite precursor alphaketoglutarate.
Major Conformational Change
Involves the flexible loop region, induced by cofactor/substrate binding. What interactions trigger the change? What parts of the loop govern substrate specificity?
3. Interface (Allosteric Regulation) explores the subunit interface in the cannonical dimer structure of MDH which plays a significant role in cooperative binding and in a “reciprocating subunit” mechanism for the enzyme. This research area is starting to detail the interactions that both trigger and mediate subunit interactions.
Is there a molecular switch at the interface responsible for subunit interactions?
4. Citrate Regulation is complex in Malate Dehydrogenases, involving both activation and inhibition depending on species and isoform. While only binding at the active site is reported, Citrate does not appear to be a competitive inhibitor. Crystal structures of inhibited and uninhibited forms show two major areas (both involving alpha helices) of the protein show significant conformational differences. What triggers these conformational changes and how they impact overall function is currently unknown
5. Loops & Turns (Protein Folding & Stability) explores the roles of the various loops and turns found in the protein and how they are involved in both structure acquisition (folding) and global and local stability. What are the rules that govern folding in acomplex alpha/beta protein such as Malate Dehydrogenase? What are the links between stability/flexibility and enzyme function?
Are there connections between folding, dynamics and allostery?
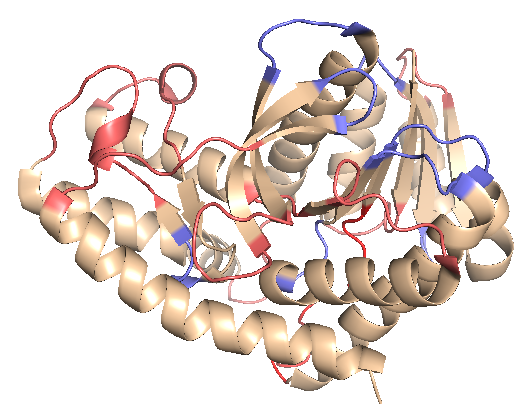
In addition to alpha helices and beta strands, malate dehydrogenase contains a number of turns (blue) and loops (pink). What roles do they play in structure function relationships?
6. Rossmann Fold (Cofactor Binding & Specificity). Like most dehydrogenases, Malate Dehydrogenase has a so-called Rossmann fold associated with confactor binding. Many questions remain including how the Rossmann fold is tailored to recognize NAD versus NADP in various isoforms, and which components of the Rossmann fold contribute to cofactor induced conformational changes involved in catalysis and in regulation.
While MDHs contain a classic Rossmann Fold involved with cofactor binding, there is an additional helix (shown in gray). What role does it play in cofactor binding or associated conformational changes?
In addition to work with Watermelon Glyoxysomal MDH, each of the above areas can involve comparative enzymology with any of the species of MDH available through the Malate Dehydrogenase CUREs Community. Three such examples are represented below.
7. Arabidopsis Malate Dehydrogenases: Organelles & Photosynthesis, Cofactor Specificity. The Arabidopsis family of Malate Dehydrogenases contains organelle specific isoforms (Mitochondrial, Glyoxysomal, Chloroplastic) as well as Cytosolic. The chloroplastic isoforms exists in both an NADPH as well as an NADH specific isoform and are subjuct to a unique redox regulation.
Chloroplastic NAD vs NADP Isoforms
The Arabidopsis NADP specific enzyme (tan) has an overall similar dimer structure and conformation as the NAD specific isoform (blue). The location of potential redox regulated disulphides in the NADP specific isoform are shown in yellow
8. Plasmodium Falciparun Malate Dehydrogenase & Drug Design. Plasmodium falciparum, like other apicomplexan forms of MDH are tetrameric, comprised of a dimer of dimers, creating several unique subunit interfaces in addition to the standard dimer interface that have been subject to potential drug design. An alternative approach to potential drug design involves the recently discovered Cryptic Allosteric site.
Combine computational and wet lab approaches to help design potential drugs for pathogenic species MDHs
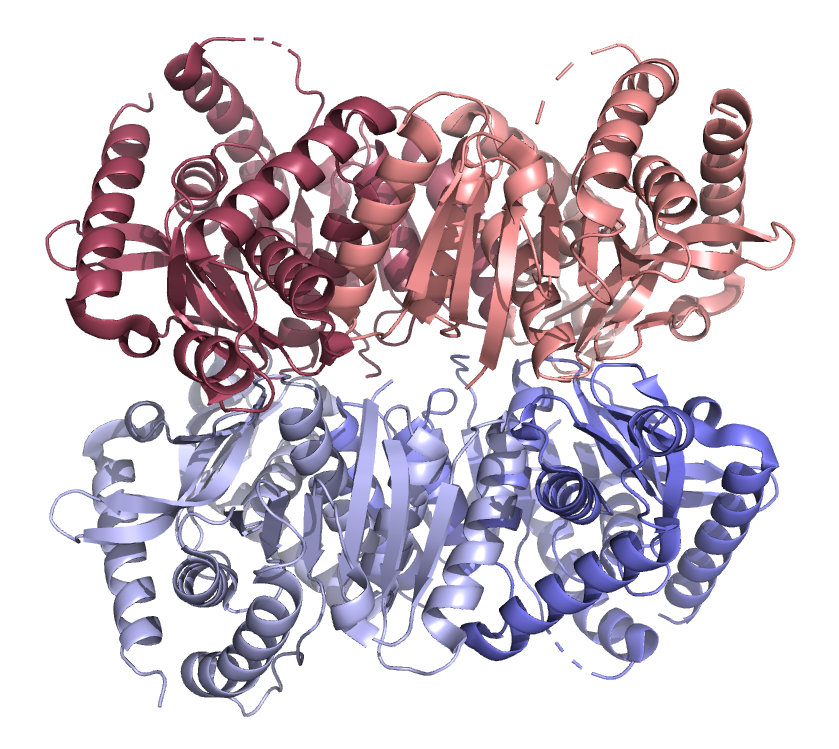
9. Ignacoccus Islandicus Malate Dehydrogenase is a primitive form of MDH (also tetrameric) showing a semblance of dual nucleotide specificity and enhanced three carbon substrate utilization. It is thought to be a bridge between Malate Dehydrogenases and Lactate Dehydrogenases.
Tetrameric Malate Dehydrogenases
such as that from Ignacoccus Islandicus off the challenge of understanding the roles of multiple interfaces as well as evolutionary relationships to other Malate Dehydrogenases and Lactate Dehydrogenase
Each of these areas involve many unanswered questions that can form the basis of an effective Malate Dehydrogenase CUREs Community project involving either a single class or collaboration between classes at different institutions.